We are grateful to the Colorado Office of Economic Development and International Trade for funding our work on developing cancer biomarkers based on cfDNA epigenomics.
News
Welcome Abby and Giovana!
We are delighted to welcome Abby Trouth (Structural Biology and Biochemistry Program) and Giovana Breda Veronezi (Molecular Biology Program) to perform their dissertation research in our lab.
Cancer and Aging Pilot Award
We thank the University of Colorado Cancer Center for awarding us a pilot grant to pursue an exciting collaboration with the DeGregori lab!
Lab’s first research paper is out!
We are excited to see the lab’s first research paper published! Please find it here: https://authors.elsevier.com/a/1cj2a3vVUPGJ0h
This paper was first posted as a preprint and Srinivas presented this story to the fragile nucleosome community sometime back.
We welcome two rotation students for Winter 2020-2021!
Abby Trouth is a graduate student in the Structural Biology Program.
Giovana Veronezi is a graduate student in the Molecular Biology Program.
Bianca wins the MCO trainee career development award
Bianca won the trainee career development award for her abstract at the Molecular and Cellular Oncology retreat at University of Colorado Cancer Center!
First Lab Publication! Review connecting cfDNA and chromatin biology
Another bright spot for the Ramachandran lab in 2020 is our first publication reviewing epigenomic methods that probe cell-free DNA in disease. Our angle, in line with the lab focus, is how chromatin structures captured in the blood can inform us about the biological state of a person.
Circulating cell-free DNA (cfDNA) provides an opportunity to develop blood tests for cancer. cfDNA harbors tissue-based information from which it originates and is of major interest for detecting cancer mutations. Surprisingly, cfDNA is generated in our bodies similar to the laboratory methods used to profile how DNA is packaged in our cells. Our review highlights connections between cfDNA features beyond the DNA sequence and the state and identities of cells that give rise to cfDNA. cfDNA provides the means to apply knowledge from the field of DNA packaging towards diagnosing and understanding cancer in humans.
Lab’s first preprint is out!!!
Check it out here: https://www.biorxiv.org/content/10.1101/2020.08.17.253146v1
Here is a tweetorial on the paper:
Excited to share the first preprint from our lab!!! We started by asking a simple question of how proteins bound enhancers and ended up with really cool results on transcription factor cooperativity. 1/
We used short (<50 bp) MNase-protected fragments to identify TF binding at high-resolution in enhancers in Drosophila cells and found on average 4 (!!!) TF binding events within ~500 bp enhancers 2/
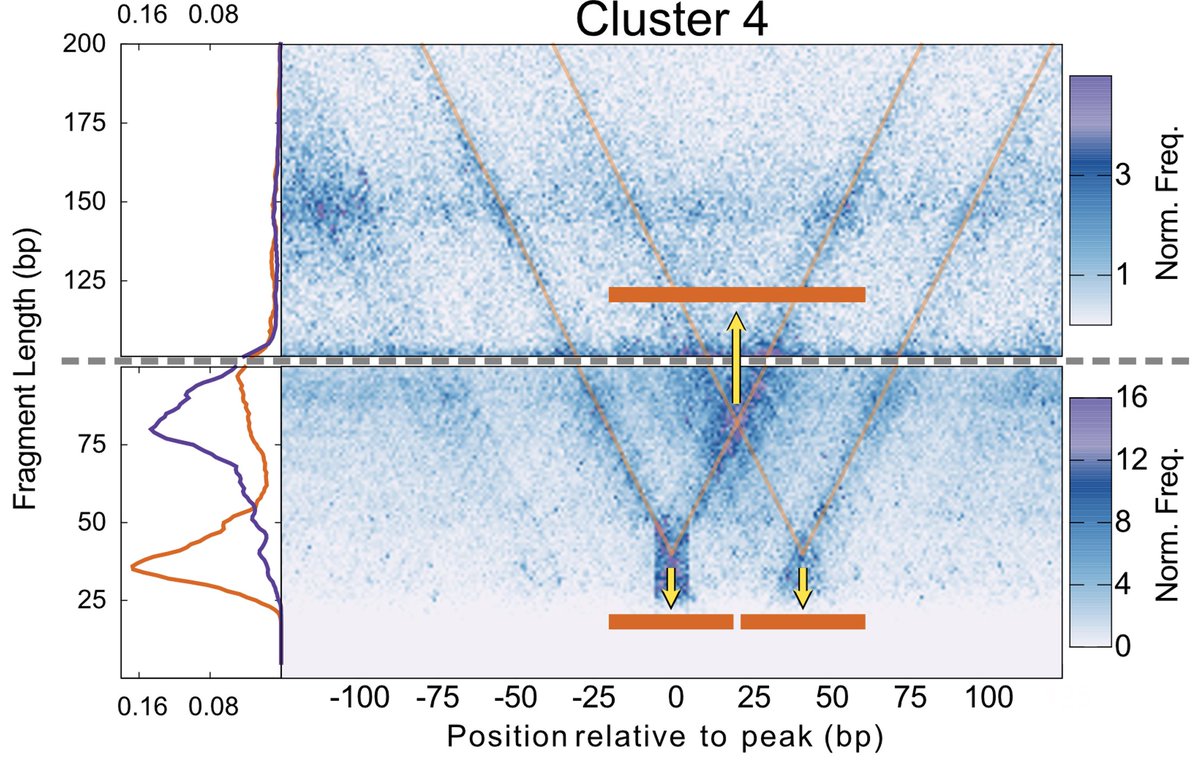
We found a way to use V-plots to ask if two of the TFs bound at the same time in these enhancers – and found most of the time they do! 3/
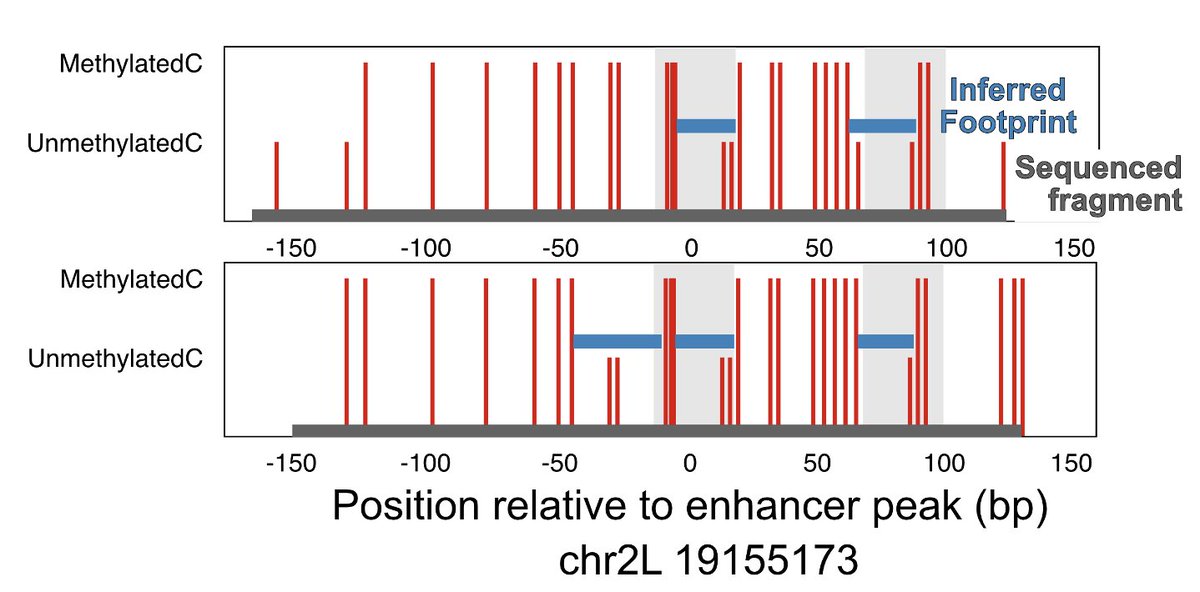
We orthogonally confirmed TFs binding at the same time using dSMF data generated by @arnaud_kr in the same cells
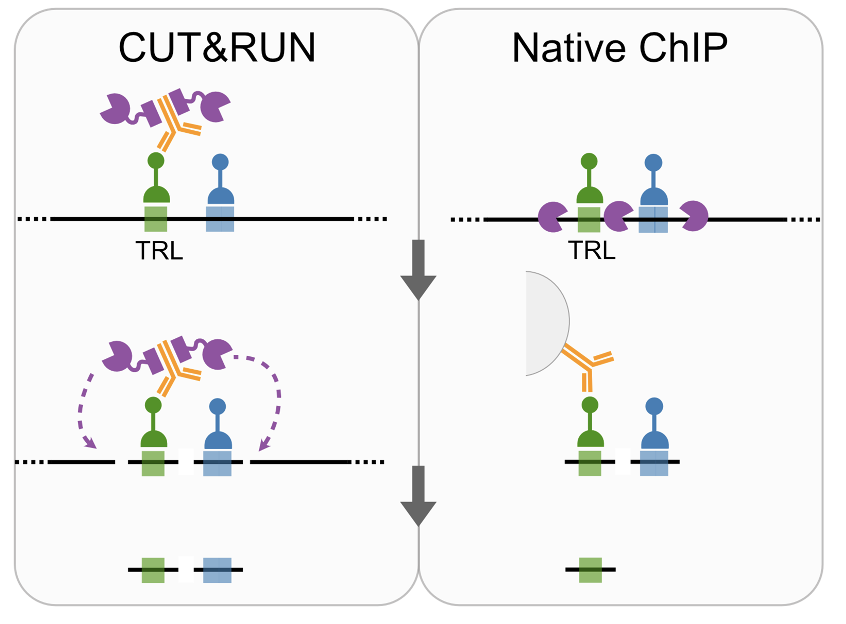
We also confirmed co-binding of TFs by comparing Native ChiP and CUT&RUN at GAGA factor (TRL) binding sites 5/
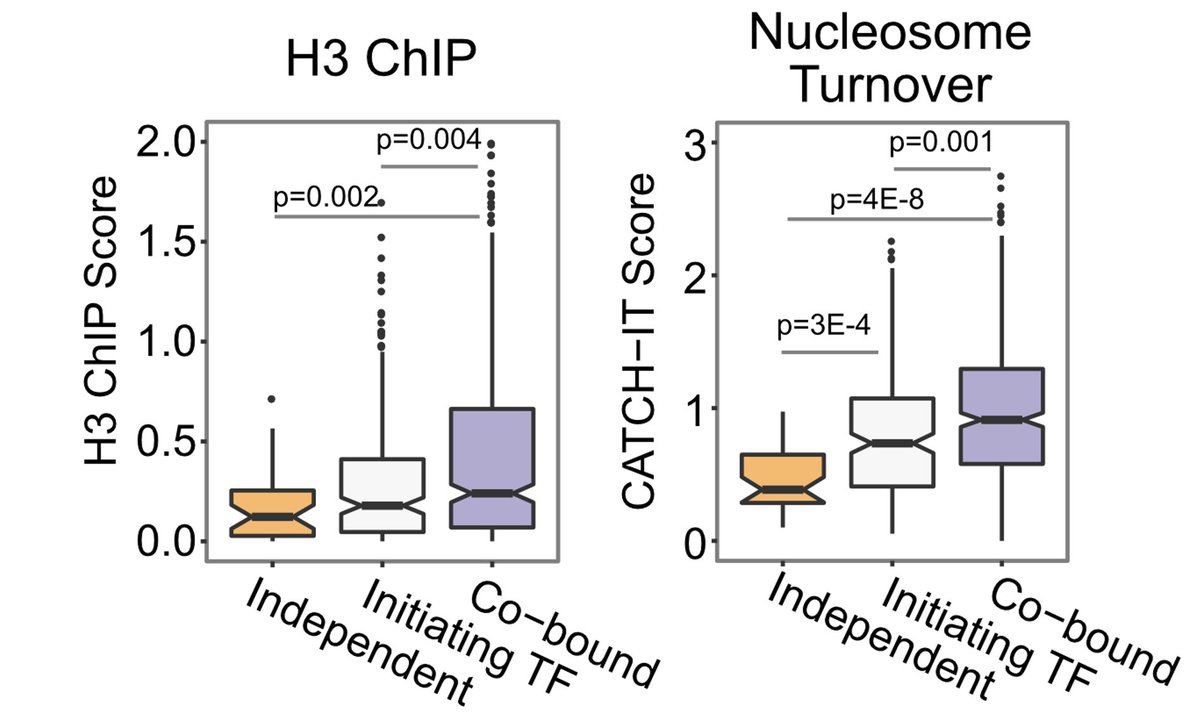
Finally, we saw cooperativity of TF binding at enhancers to correlate with BOTH nucleosome occupancy and turnover, pointing to cooperative binding as a means to displace nucleosomes from enhancers 6/
I want to thank my co-authors and collaboratorsr @satyaiiitm @KamiAhmad3. And this work wouldn't have been complete without amazing previous work from @AlexanderStark8 on STARR-seq and @arnaud_kr on dSMF!
Originally tweeted by Srinivas Ramachandran (@4everBiochemist) on August 18, 2020.
Srinivas presents at Fragile Nucleosome seminar series
Check out his talk on youtube:
Alexis wins the CRTEC Innovation award
Alexis won the CRTEC innovation award from University of Colorado Cancer Center!